About EpiGePT
EpiGePT, a transformer-based model for cross-cell-line prediction of chromatin states by taking long DNA sequence and transcription factor profile as inputs. With EpiGePT, we can investigate the problem of how the trans-regulatory factors (e.g., TFs) regulate target gene by interacting with the cis-regulatory elements and further lead to the changes in chromatin states. Given the expression profile of hundreds of TFs from a cellular context, EpiGePT is able to predict the genome-wide chromatin states given the cellular context.
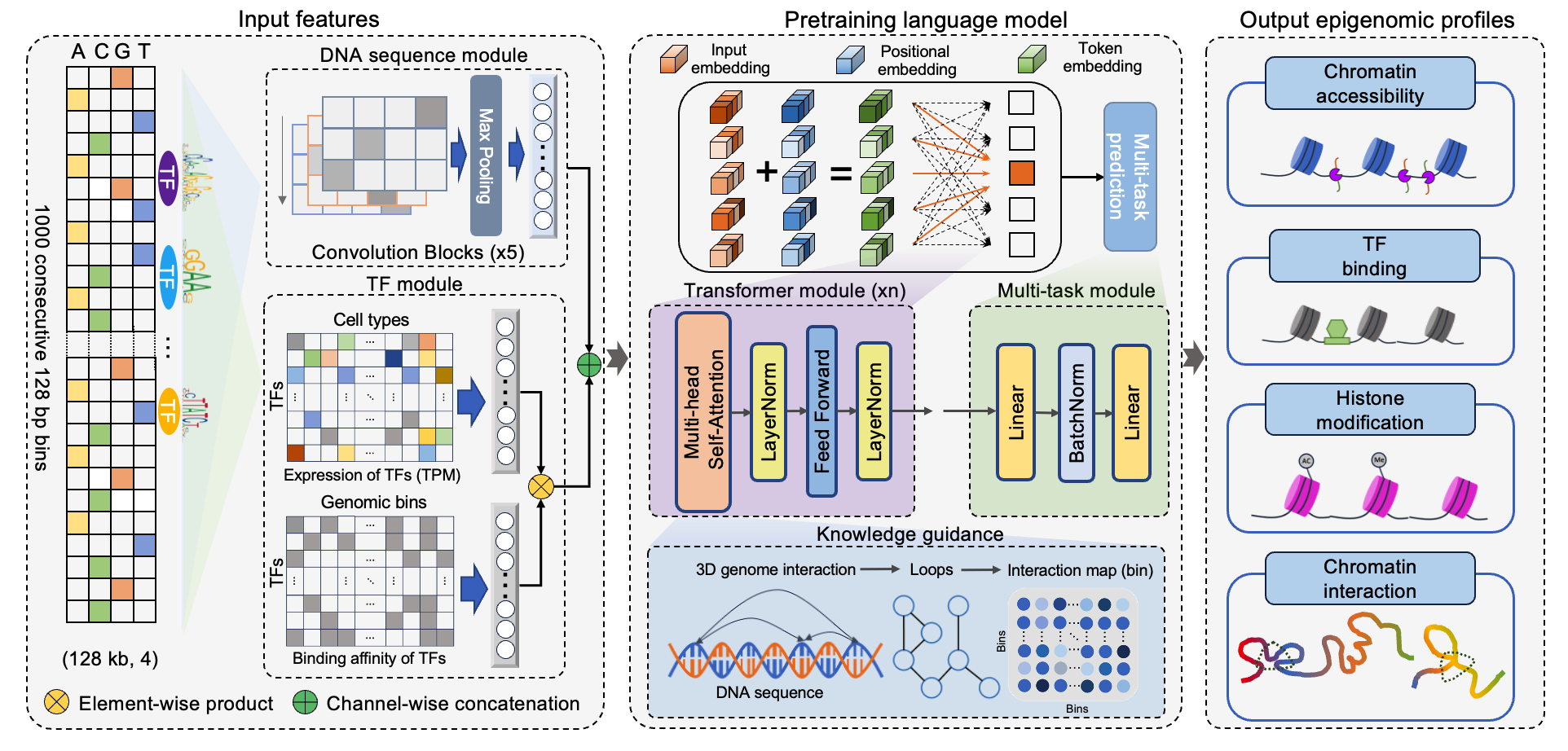
You can obtain the prediction of genomic feature of specific genomic regions (hg19 and hg38) including DNase, CTCF, H3K27ac, H3K4me3, H3K36me3, H3K27me3, H3K9me3 and H3K4me1 by submitting a BED file to this web server.
When you select a locus, we provide predicted values of epigenomic signals for a region upstream and downstream of the locus, spanning 128 kilobase pairs (kbp). The region is divided into 1000 genomic bins, each measuring 128 base pairs (bp), and the predicted values for each bin are provided.