The documentation of collectNET
Home page
- Brief introduction to collectNET: What is collectNET, why we developed such a tool, and its main applications.
- Brief introduction and flowchart of online cell communication inference and analysis, along with a quick access link to the analysis page.
- Overview of the human organ reference atlas, including links to browse interfaces for each organ, statistics on the number of cell types in each organ, and statistics on the number of significant interactions they contain.
- Links to relevant databases for collectNET applications.
- News and updates about collectNET.
The home page primarily provides an overview of collectNET, quick links to access online services and browsing, and other necessary information.
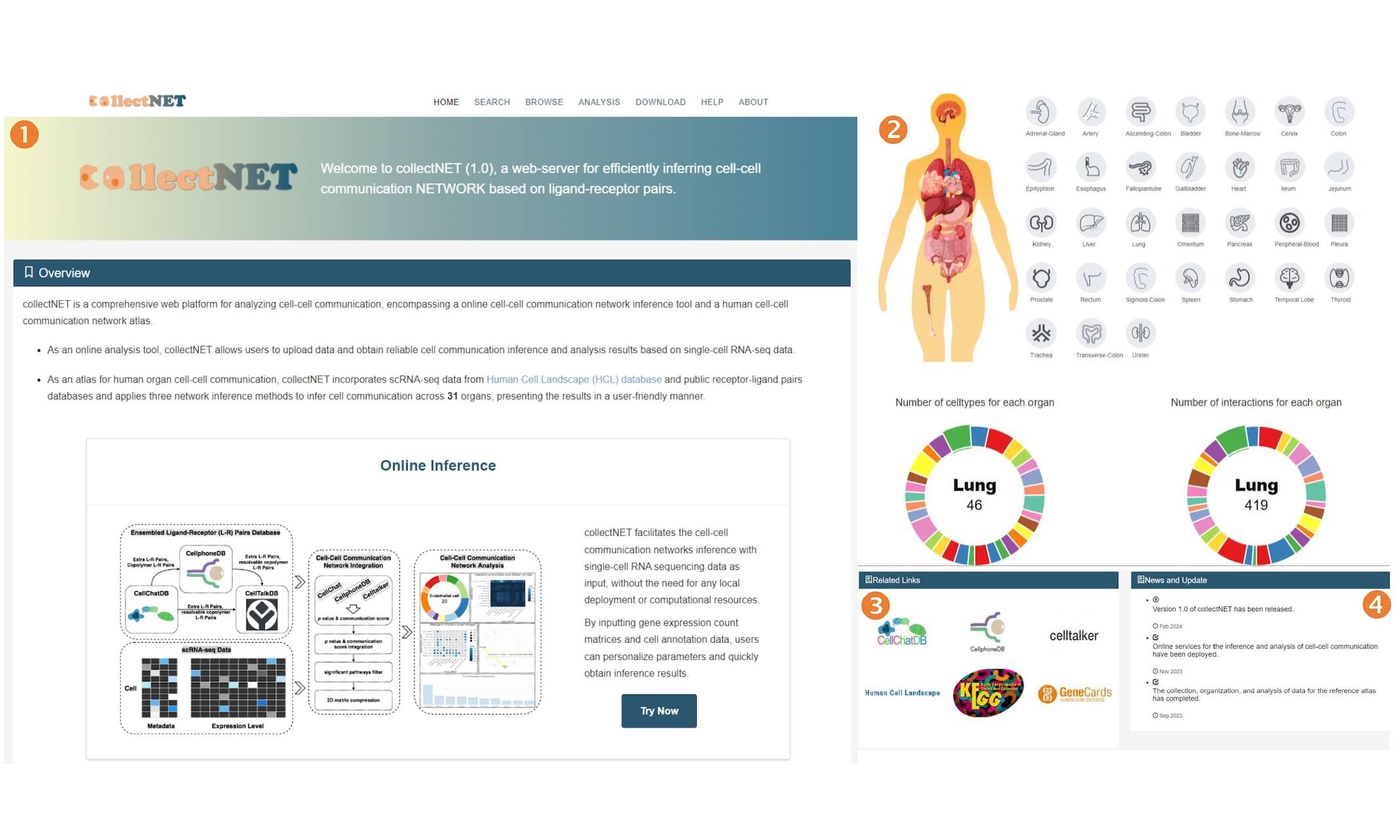
Search page
- In the search section, users can input or select different methods, organs, cells, receptors, interactions, and sort the output results by p-value or communication score. Clicking the example button will provide a sample search, and clicking the submit button will display the output results.
- In the results section, we present the search results in a table format. To provide more comprehensive information, we provide the KO number and link to the receptor in the KEGG database. For ligands or receptors not included in KEGG, we provide a GeneCards link in the ligand details column. Additionally, users can download their search results in table format using the Download button.
Users can conveniently search all data in our reference network atlas based on specific criteria using the search page.
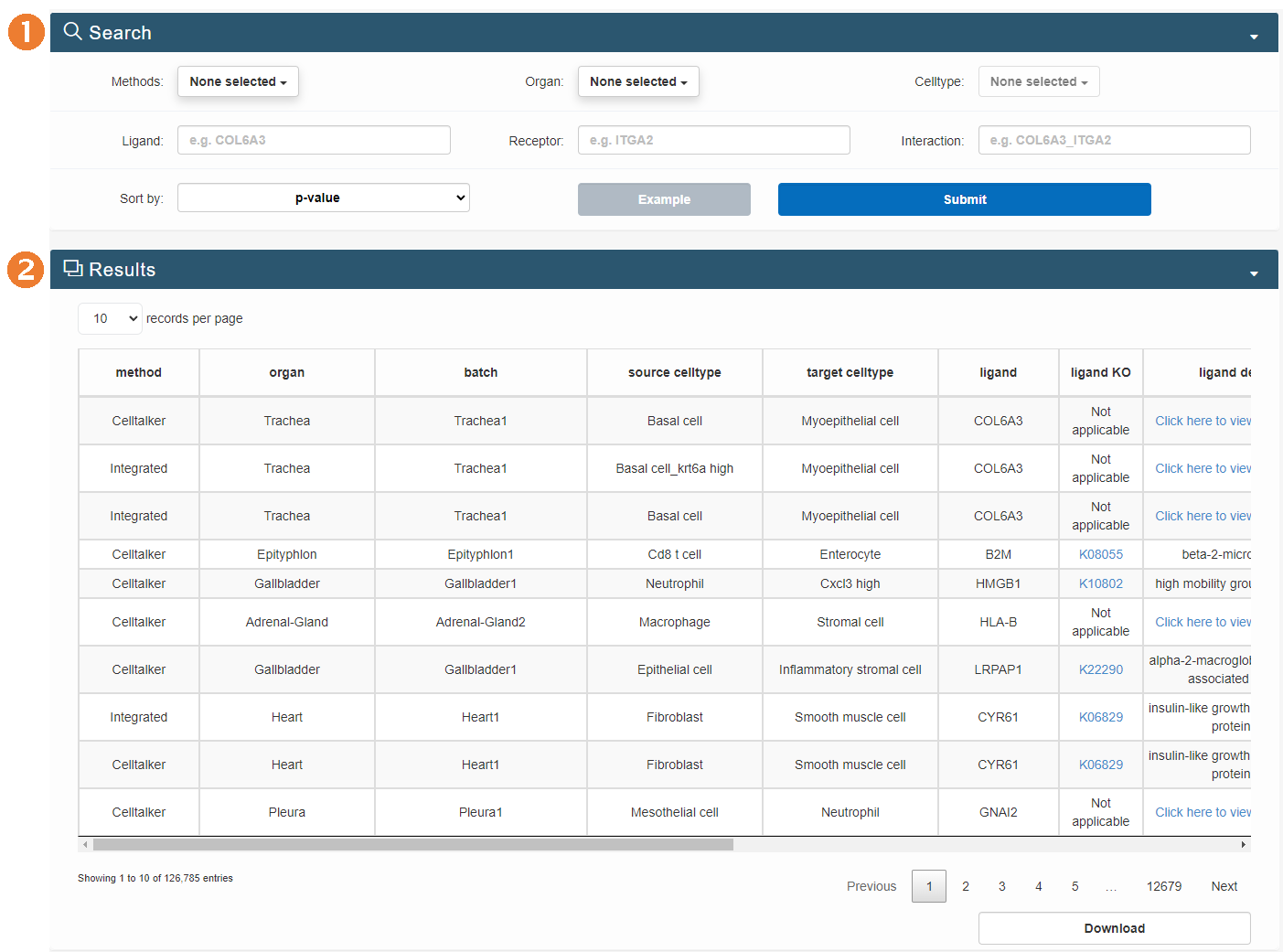
Browse page
- Clicking on organ names will display statistical graphs of cell types within that organ, the top 10 most common ligands and receptors, and a graph of receptor-ligand interactions. The table below will show all significant receptor-ligand pair information for that organ.
- Some organs have multiple batches of data. Clicking on the batch names will display the UMAP plot, circular plots for three inference methods, and an integrated method. The table below will show all significant receptor-ligand pair information for that organ batch.
The browse page mainly includes browsing the organ-specific and batch-specific reference atlas.
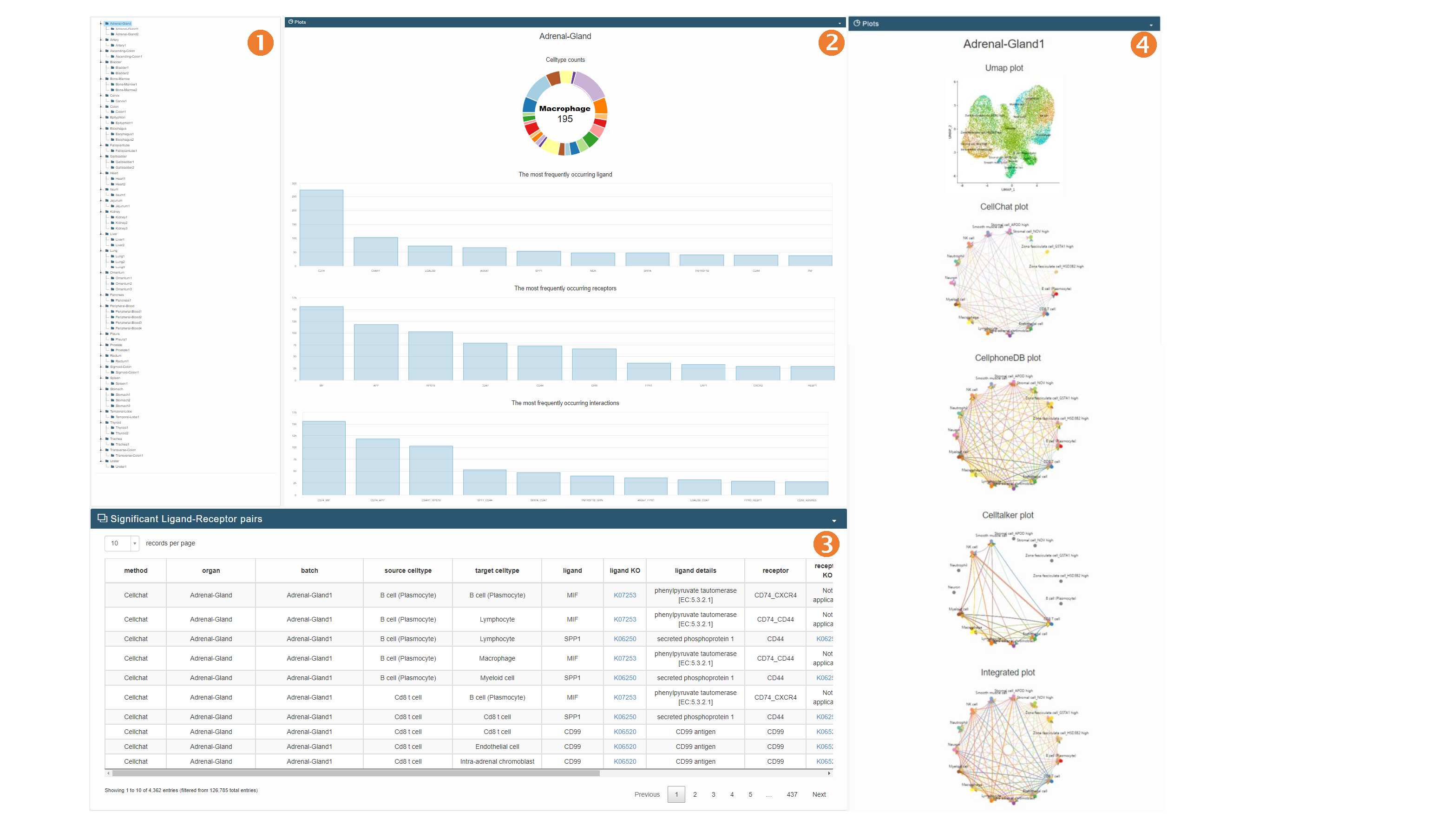
Analysis page
- Supports user-uploaded single-cell RNA-seq data: To facilitate data organization for users, we read in cell-by-gene matrices from txt files, where the row names and column names correspond to the cell names and gene names, respectively. Additionally, we require a cell annotation file in csv format, where the row names are cell names that correspond one-to-one with the cell names in the previous txt file, and the column names containing cell annotation information should be named "Celltype". Here, we provide an example, and you can click on "example" to view the corresponding input data example.
In this interface, users can perform cell communication online inference and analysis using collectNET, which is the main feature of our online analysis tool. Input Interface
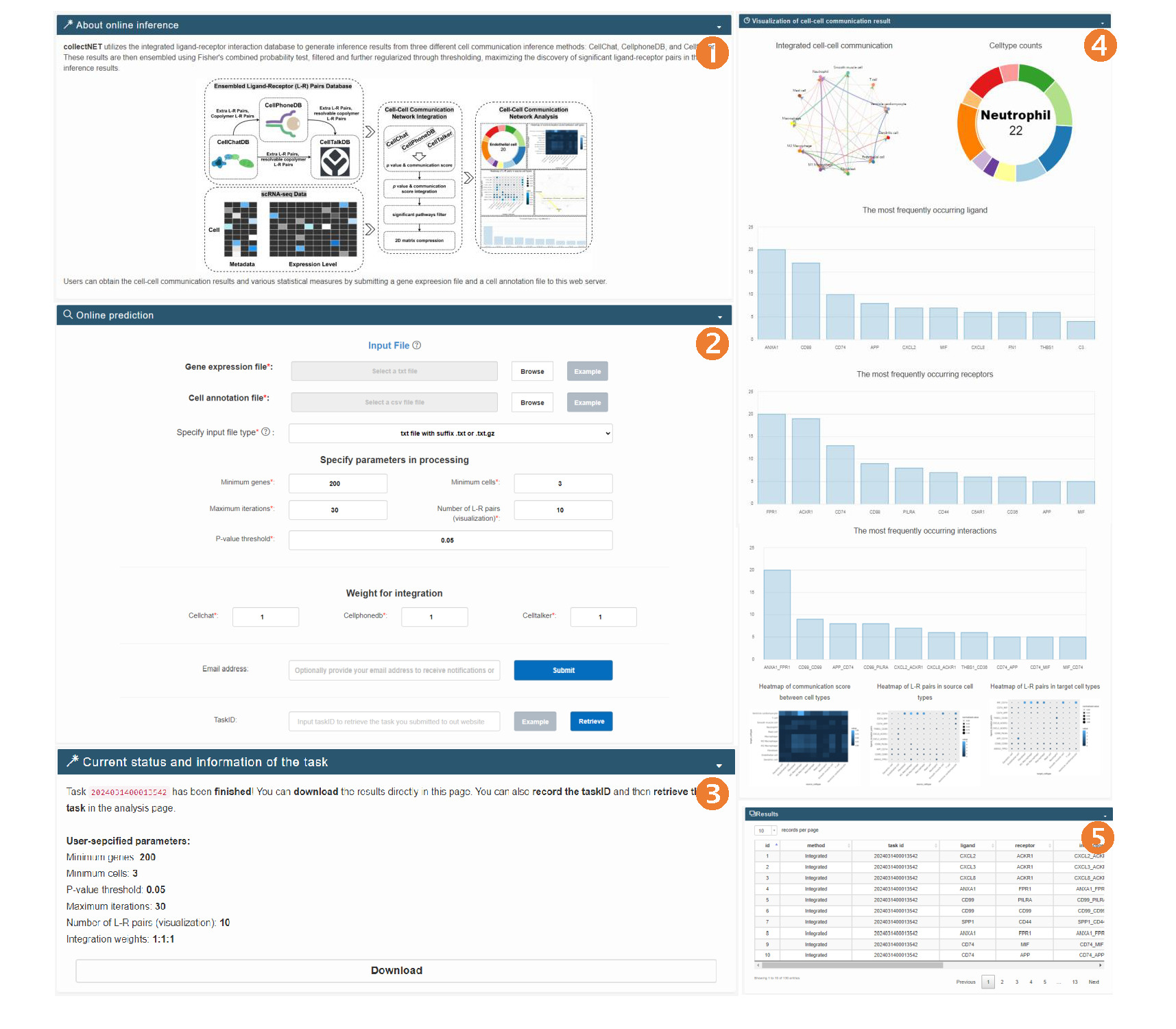
- Before online analysis, users can select input parameters. The "Minimum genes" and "Minimum cells" are parameters for data preprocessing, representing the minimum number of expressed genes in a single cell and the minimum number of cells in which a single gene is expressed, respectively. The "p-value threshold" represents the threshold for significant p-values in the integration of receptor-ligand pairs, where values below this threshold are considered significant receptor-ligand pairs.
- If your data is large, running the analysis may take up to a dozen minutes. We recommend leaving your email address to conveniently receive the inference and analysis results.
- After submitting the input file and setting parameters, users can obtain the current status and information according to the taskID. Outputs the current status, parameters, and an output table containing information on significant receptor-ligand pairs during the running process.
- In the visualization section, an integrated cell communication circular plot is displayed, along with cell type information in the dataset, the top 10 most frequently occurring ligands and receptors, and bar plots and heatmaps of receptor-ligand pair interactions. In the table below, all significant receptor-ligand pair information in the dataset will be displayed.
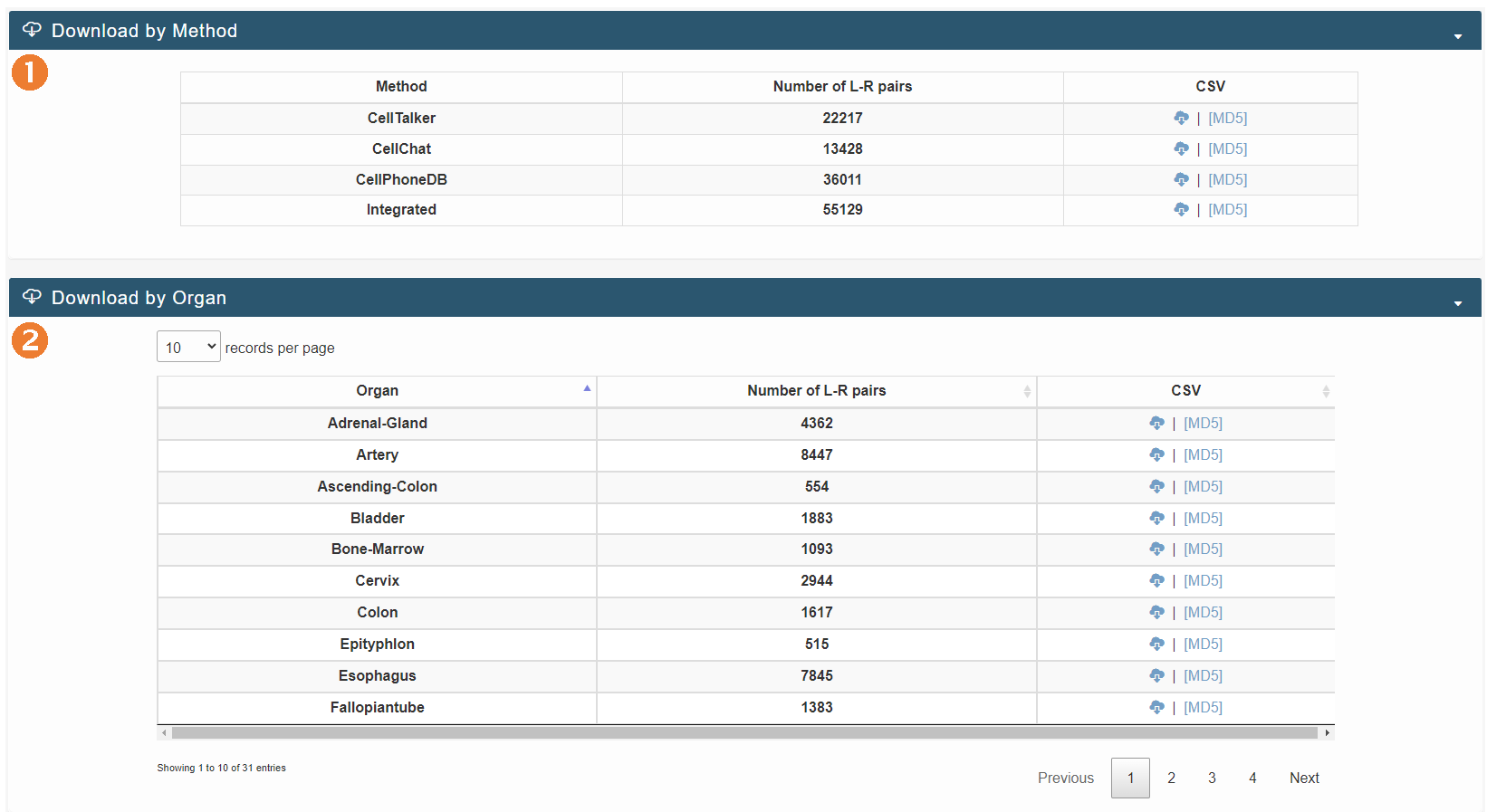
Download page
-
The download interface allows users to download data from the human organ cell communication atlas provided by us, based on the inference methods and organs.
About page
-
The about page provides information about the authors of collectNET and their contact details. If you have any questions or inquiries regarding collectNET, please feel free to contact us at any time.
The browser compatibility of collectNET
| Operating System | Version | Chrome | Firefox | Opera | Microsoft Edge | Safari |
|---|---|---|---|---|---|---|
| Windows | Windows 10 Pro | |||||
| MacOS | Sierra 10.12 | Linux | Ubuntu 16.04 |
Frequently asked questions
What is collectNET?
collectNET, to the best of our knowledge, is the first web-server for online integrated inference of cell-cell communication. collectNET has constructed a cell communication atlas of the human single-cell database, including cell communication of 31 organs, 485 cell types, and 126,785 significant ligand-receptor pairs. With user-friendly functionalities, efficient calculation, hierarchical browsing, comprehensive statistical analysis, advanced search capabilities, and intuitive visualization, collectNET shows numerous advantages compared to existing tools.
What features collectNET?
- collectNET integrates CellChat, CellPhoneDB, and CellTalker using Fisher's combined probability test, providing more reliable communication pathways.
- collectNET integrates three widely-used databases for ligand-receptor pairs as prior knowledge, enabling more comprehensive analyses.
- collectNET furnishes a more extensive cell communication resource, encompassing 31 organs, 485 cell types, and 126,785 communication pairs of human organ.
- collectNET enables users lacking coding expertise or computational resources to perform efficient cell communication inference and visualization through intuitive tutorial-style interfaces.
- collectNET offers downloadable tables for different methods, organs, and user-submitted inference tasks, providing a rich foundation for broad applications, such as drug target prediction, tumor microenvironment analysis, and immunotherapy strategies.
Why are the advantages of collectNET compared with other tools?
- It is true that several methods such as CellPhoneDB (Nature Protocols, 2020), CellTalker (Immunity, 2020), and CellChat (Nature Communications, 2021) were developed to infer cell-cell communication utilizing single-cell RNA-seq data. Nonetheless, previous studies (Nature Communications, 2022) have underscored the notable biases and lack of consistency among the inferences of these methods. Integrating the inferences from various methods necessitates complex computational workflows, time-consuming setup of the environment, and substantial computational resources. collectNET addresses this issue to some extent.
- Other database-based tools like CellCommuNet (NAR, 2023) primarily focus on known inference results and perform comparative analysis with their own databases, but they are unable to infer cell communication from raw single-cell sequencing data. collectNET offers a comprehensive inference and analysis pipeline starting from single-cell RNA-seq data, eliminating the need for researchers to configure different environments or switch platforms.
- With prior knowledge of 3,954 ligand-receptor pairs from three public databases, systematic integration of CellPhoneDB, CellTalker, and CellChat, collectNET can provide accurate and reliable online inference service, saving users at least 2 hours for implementation.
- collectNET performs pre-processing, inference, integration and visualization. The output includes diverse statistical measures, visualization of the inference results, and a downloadable table for integrated cell-cell communication scores.
- The existing environment setup for cell communication inference tools is time-consuming and requires high computational resources. collectNET provides an efficient and convenient analysis platform for users who face challenges in both of these aspects.

How to prepare data for collectNET?
-
Here is a detailed tutorial to help you prepare input files. Click here!