The documentation of Human-scATAC-Corpus website
Home page
- A simple introduction about the database, with a global view of statistics of the scATAC-seq data we collected.
- The number distribution of cells across various tasks and tissues.
- The number distribution of data type.
- Recent articles about scATAC-seq data analysis computational methods.
- News and updates of Human-scATAC-Corpus.
- Links of relevant single-cell databases.
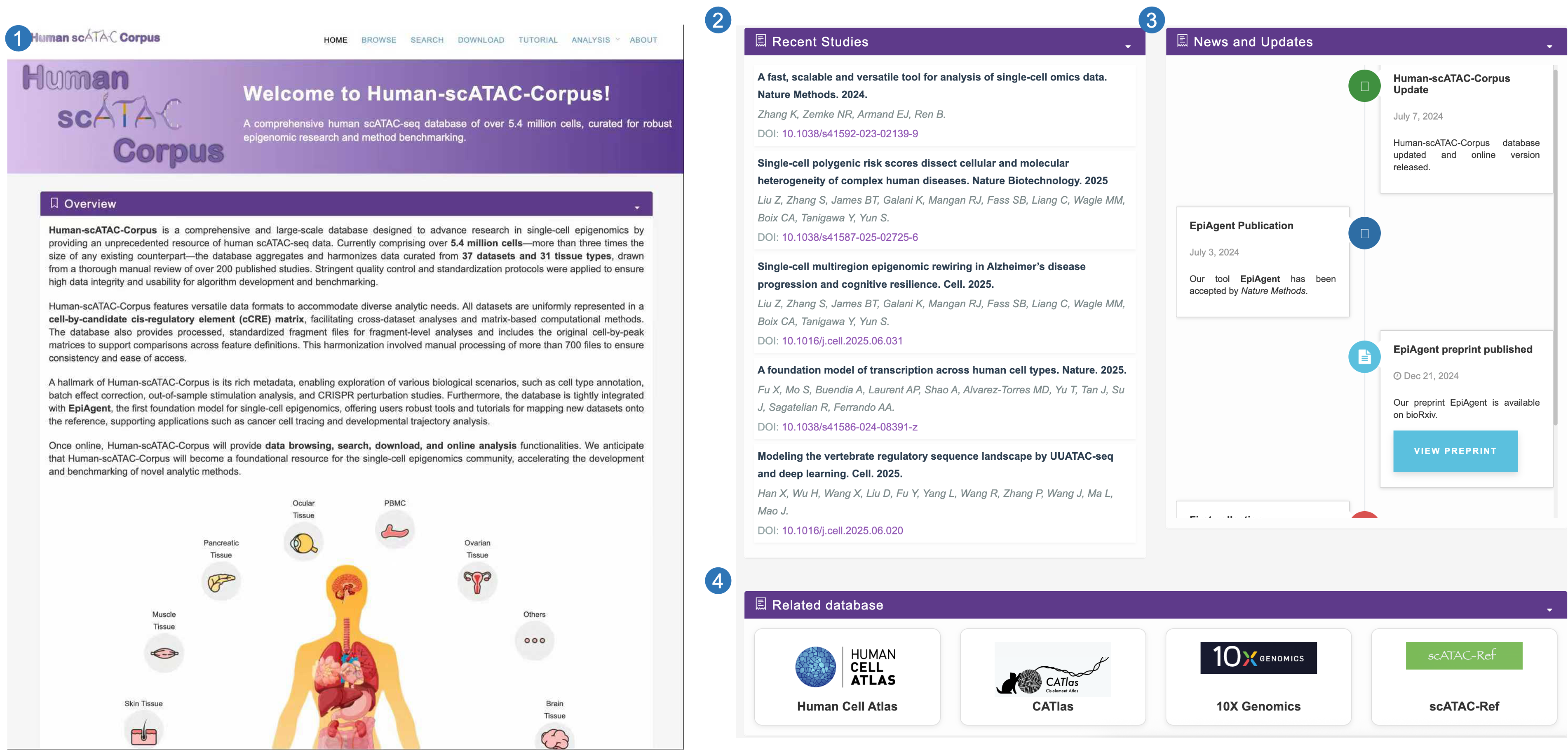
Browse page
- Users can browse samples of interest by selecting a category.
- Users can select items from different tasks, tissues, and datasets to browse the corresponding samples.
- The table presents the metadata of each sample, enabling users to download the corresponding files or click the sample ID to navigate to the detailed page.
- We provide umap visualization for the cells in each category.
- We provide comprehensive statistics with respect to the selected subset of cells.
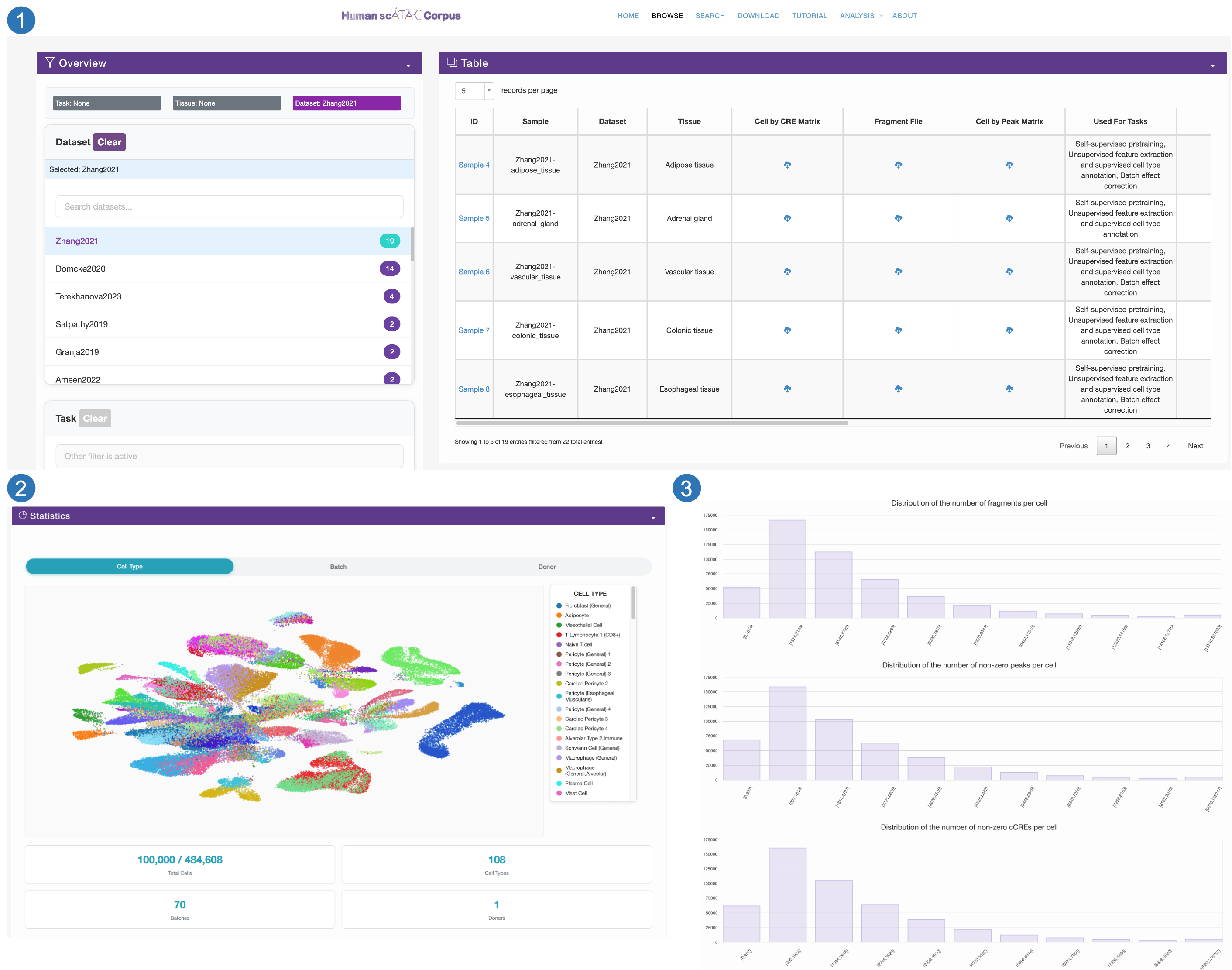
Details page
- Detailed information includes Sample,Dataset,Tissue,Used For Tasks,Availability,Raw Data Type,Corresponding Study,Number of Cells,Number of Cell Types,Number of Batches,Organ,Sequencing Technology for each sample.
- We provide the umap visualization for cells with the sample
- We also provide distribution plots of the non-zero cCRE peaks and fragment counts across different cells.
- We also provide pie charts of the different cell types, donors, and batches within the sample..
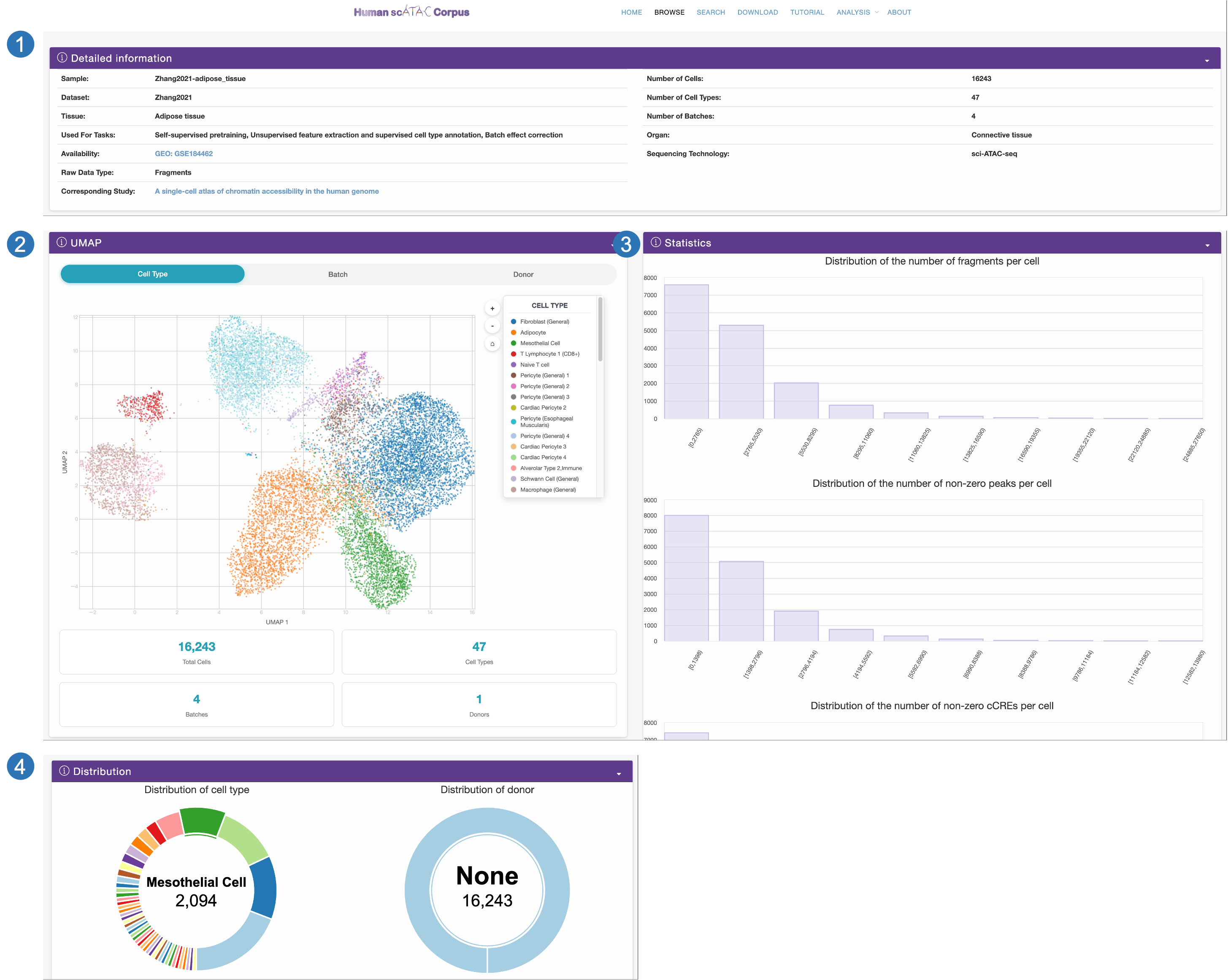
Search page
- Users can search for scATAC-seq data (sample) of interest by selecting Dataset, Tissue, Organ, Used for task, Sequencing technology, the ranges of cell counts, cell types, and batches. We also provide an example to facilitate the usage of searching.
- The results will be displayed on an interactive table.
- Users can click the ID to access detailed information about this sample on a new page.
- By clicking the Download button, users can obtain the compressed table about the metadata and download link.
Analysis page
-
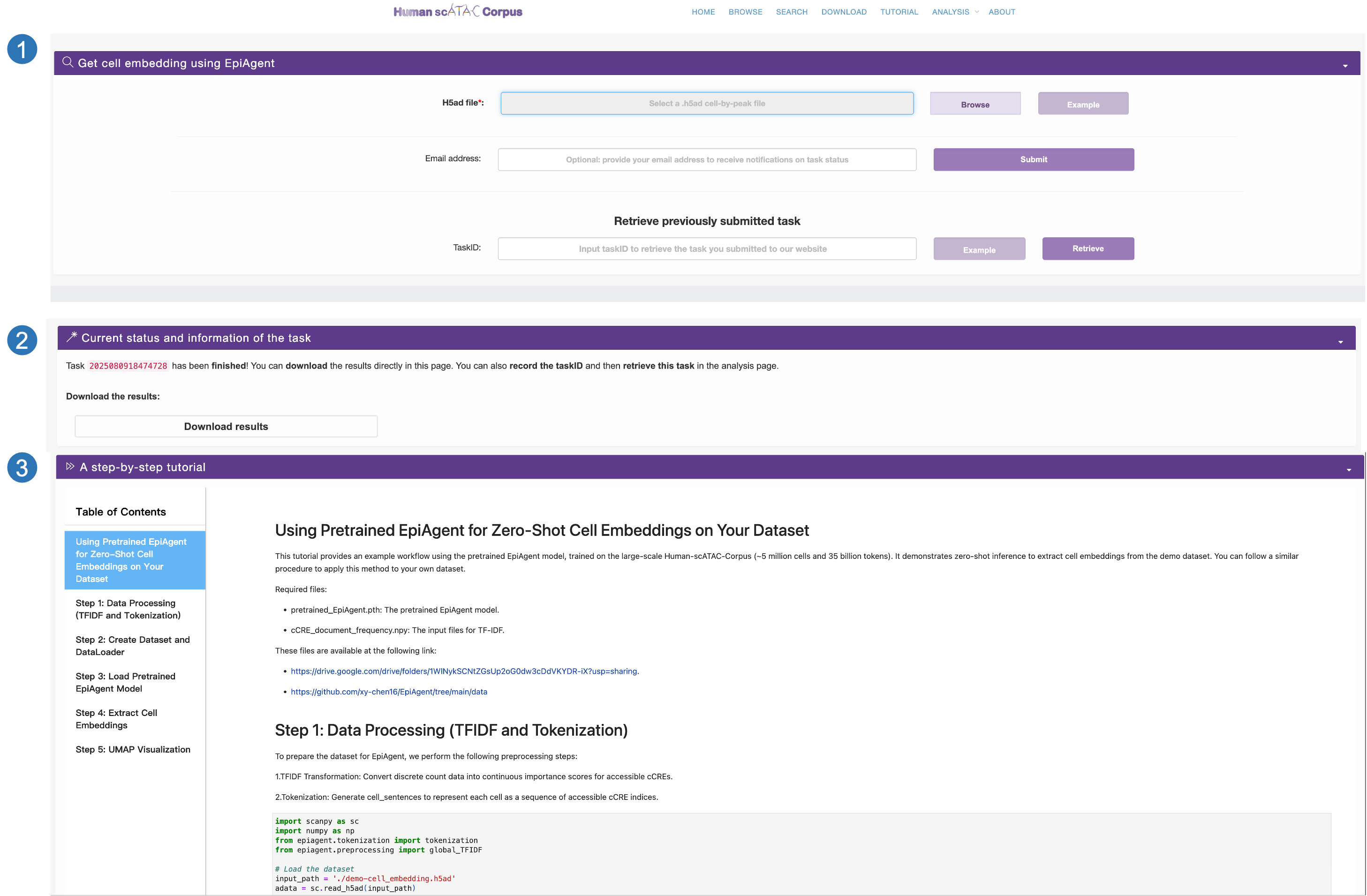
We offer three distinct online inference task services: cell embedding extraction, cell type annotation, and data mapping:
- Taking cell embedding task as an example, users are required to first select and upload their own cell-by-cCRE file and may optionally provide their email address to avoid prolonged waiting times. Users can click the Example button to download a sample file.
- Taking cell embedding as an example, users are required to first select and upload their own cell-by-cCRE file and may optionally provide their email address to avoid prolonged waiting times. Upon submission, users will receive a unique task ID for subsequent tracking. Users can click the Example button to download a sample file.
- We also provide a detailed step-by-step tutorial to guide users in applying our pretrained model locally to accomplish each of these three tasks.
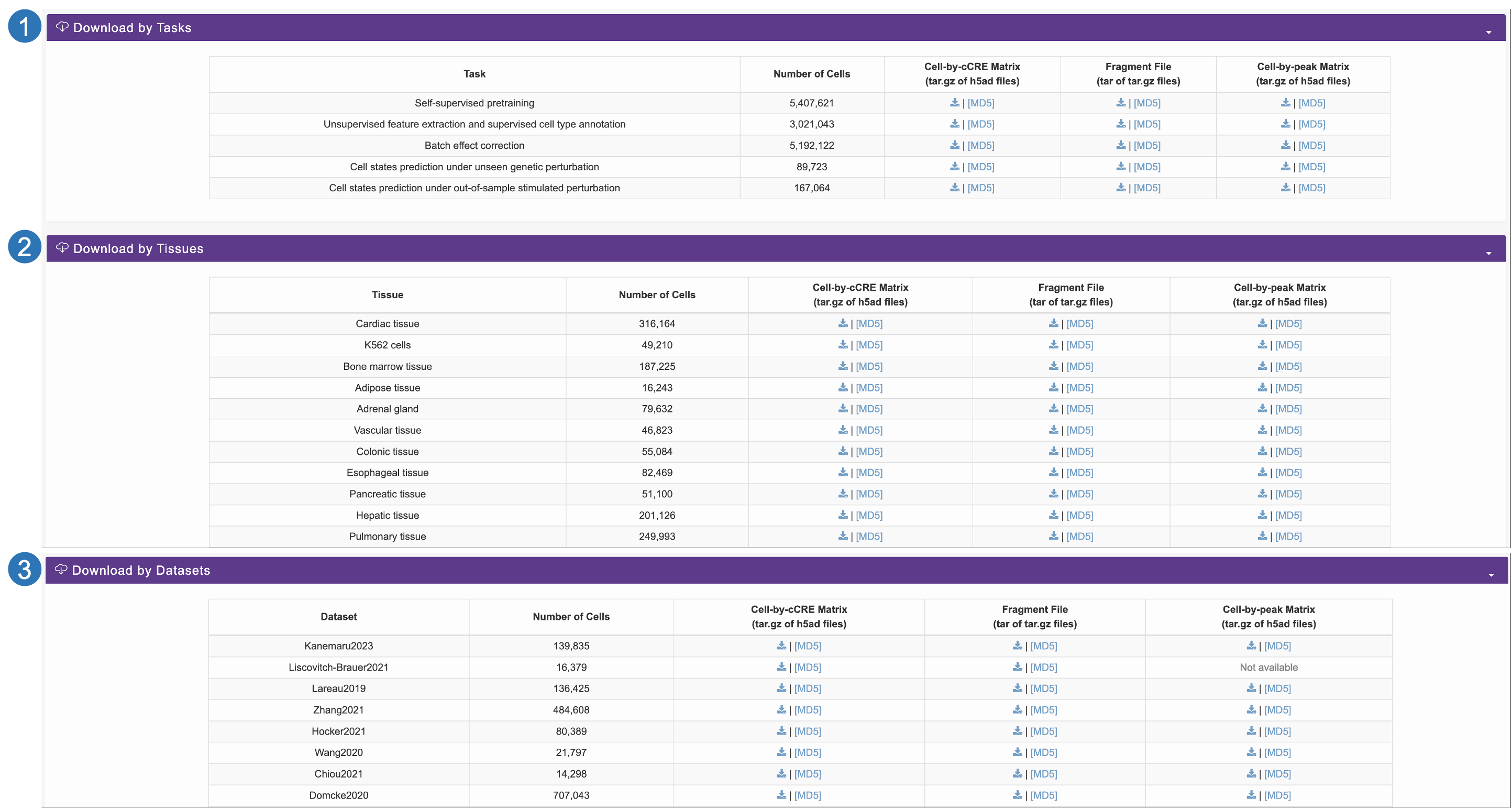
Download page
- We categorize these samples by task and provide users with downloadable files including the cell-by-cCRE matrix fragment file, the cell-by-peak matrix, and metadata.
- We also categorize these samples by tissue and dataset, and provide users with downloadable files including the cell-by-cCRE matrix fragment file, the cell-by-peak matrix, and metadata.
- The corresponding MD5 files are provided to ensure the integrity of the downloaded data.
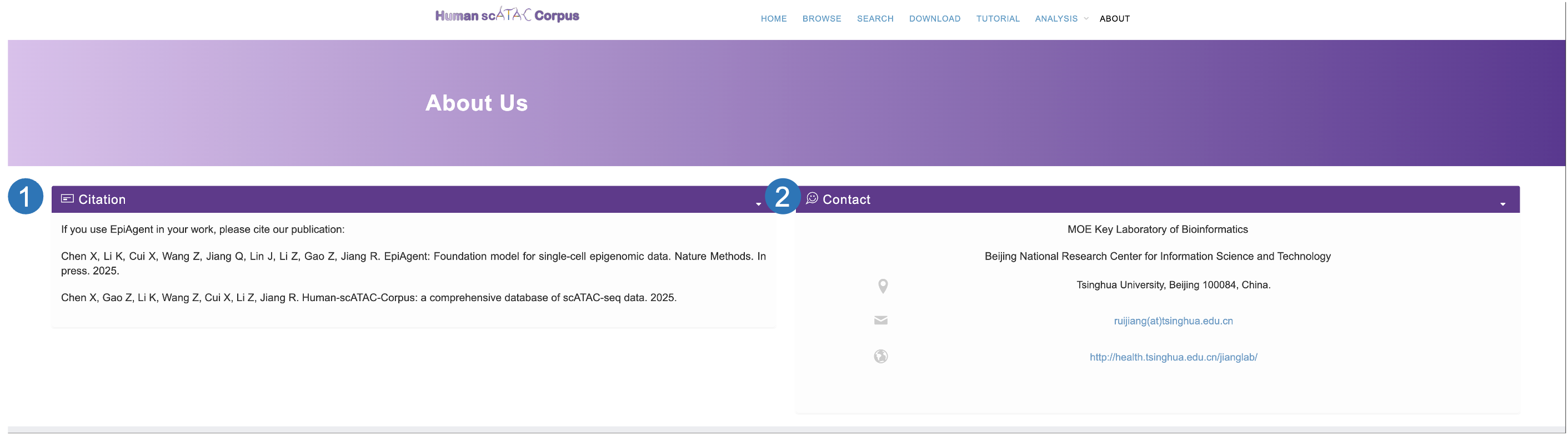
About page
- The citation information of Human-scATAC-Corpus, If you have downloaded or used data from the Human-scATAC-Corpus or its online analysis services, please cite our website as well as the EpiAgent (10.1101/2024.12.19.629312).
- The contact information of develop team of Human-scATAC-Corpus. Please feel free to contact us if you have any question about data or online service of Human-scATAC-Corpus.
The browser compatibility of Human-scATAC-Corpus
| Operating System | Version | Chrome | Firefox | Opera | Microsoft Edge | Safari |
|---|---|---|---|---|---|---|
| Windows | Windows 11 | |||||
| MacOS | Sequoia 15.5 | Linux | Ubuntu 22.04 |