The documentation of scEpiTools
Home page
- A simple introduction about the database, with a global view of statistics of the single-cell epigenomic tools we collected.
- The number distribution of tools across various methodologies, published date and main categories.
- The number distribution of published or preprint tools from 2015 to 2023.
- Recent articles about single-cell epigentic tools.
- Links of relevant single-cell databases.
- News and updates of scEpiTools.

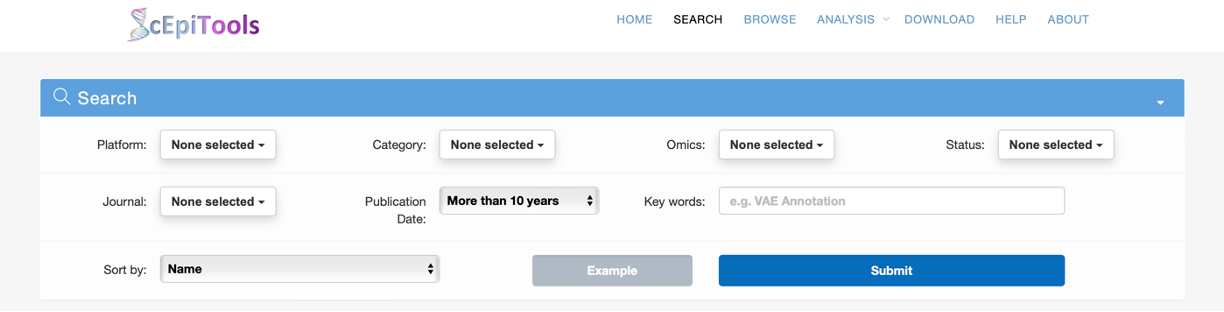
Search page
- Users can search for software tools of interest by selecting multiple platforms from the pull-down menu, and then selecting multiple categories, omics, status and journals from the pull-down menu with advanced features such as searching and selecting-all. Users can also determine the scope of the tools query by determining the publication date and key word of the tool. Users can also sort the results according to different keywords according to their needs. We also provide an example to facilitate the usage of searching.
- The results will be displayed on an interactive table.
- Users can click the ID to access detailed information about this software tool on a new page.
- By clicking the Download button, users can obtain the compressed results .

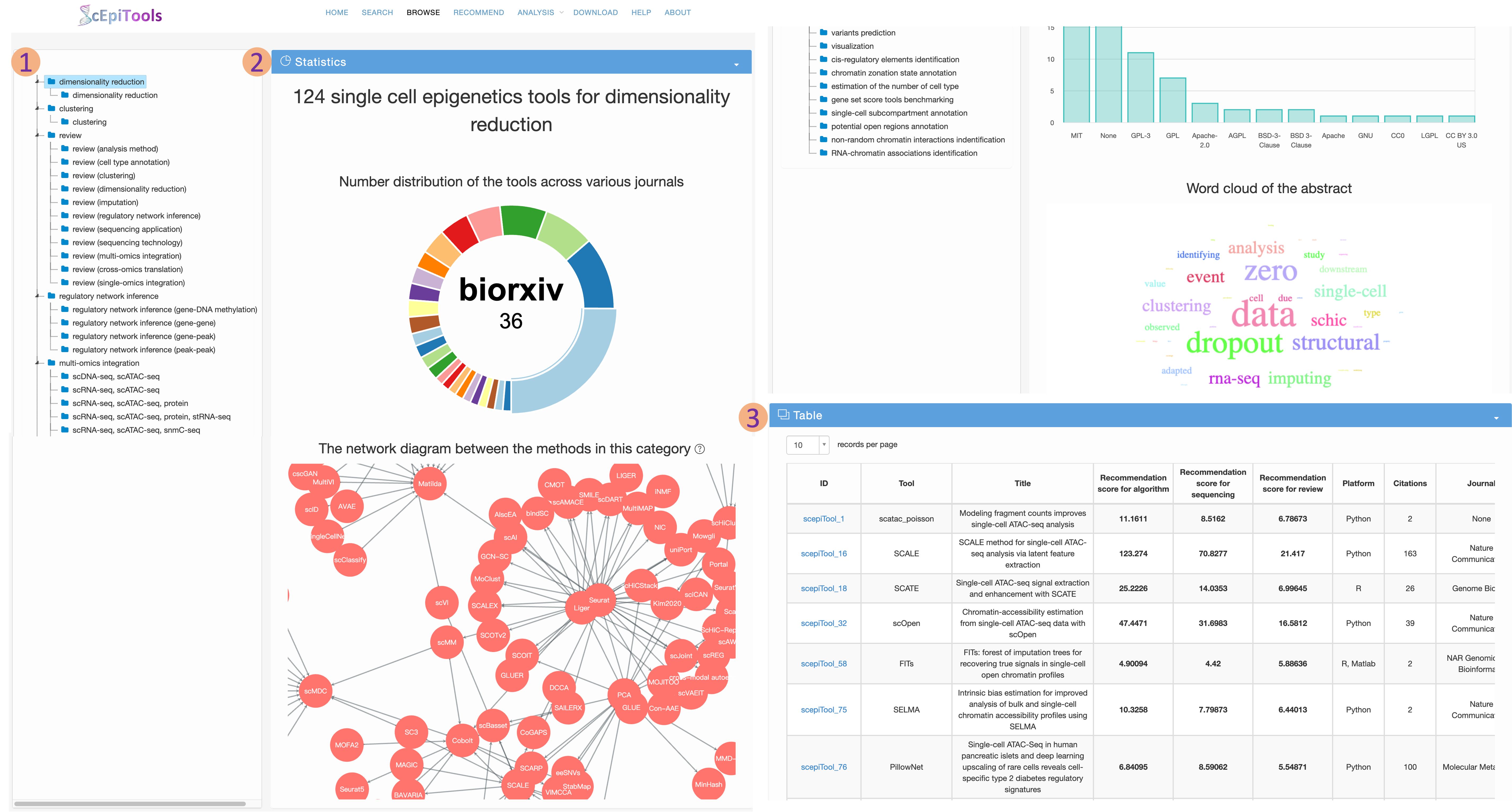
Browse page
- Users can browse tools of interest by selecting a category in the hierarchical tree.
- We provide comprehensive statistics and visualization with respect to the selected subset of software tools.
- The number distribution of tools across various main categories and sub categories
- The number distribution of the tools across various journals.
- The number distribution of the tools across various methodologies.
- The number distribution of the tools in diverse intervals of citations.
- The number distribution of the tools in different platforms.
- The word cloud of the description.
- The selected subset of software tools are displayed on a interactive table. Users can click the ID to access detailed information about this software tool on a new page.
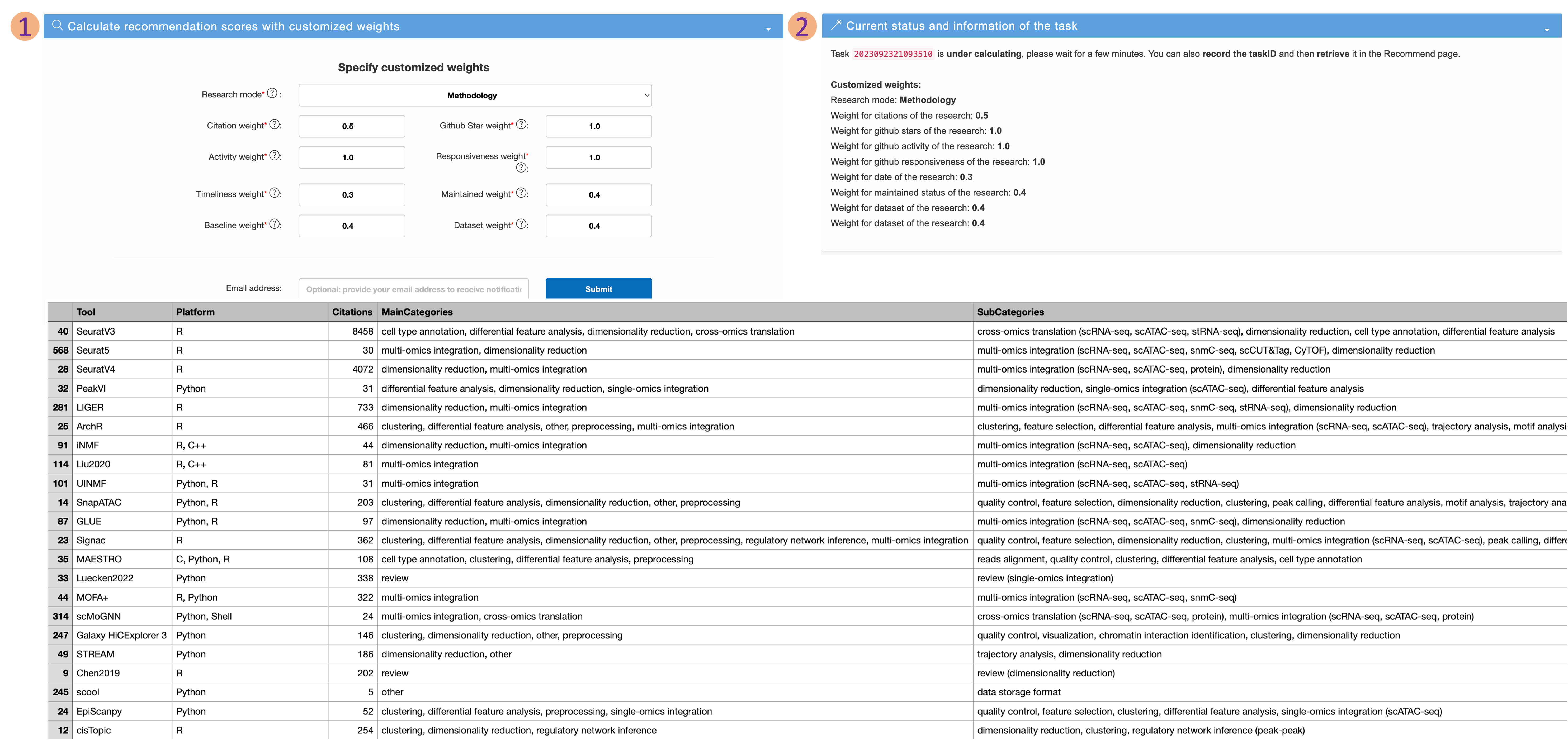
Recommend page
- Users can customize the recommendation scores by adjusting the weights of various features and submit the task. Then users can obtain the corresponding recommendation scores and detailed annotation of articles.
- Users need to first select the research mode they wish to use for recommendations, further adjust the weights of different features, and then click "Submit" button. This prompts the server to begin calculating the recommendation scores for each article in the background, according to the user's settings (see more details in the supplementary text S4 of our paper).
- Then users can retrieve the status of their tasks. Once the calculations are completed, users can click the download button to obtain a file containing article annotations and recommendation scores.
- Users can also provide their email address. Once the task is finished, scEpiTools will automatically send you an email to alert you that your task has been completed.
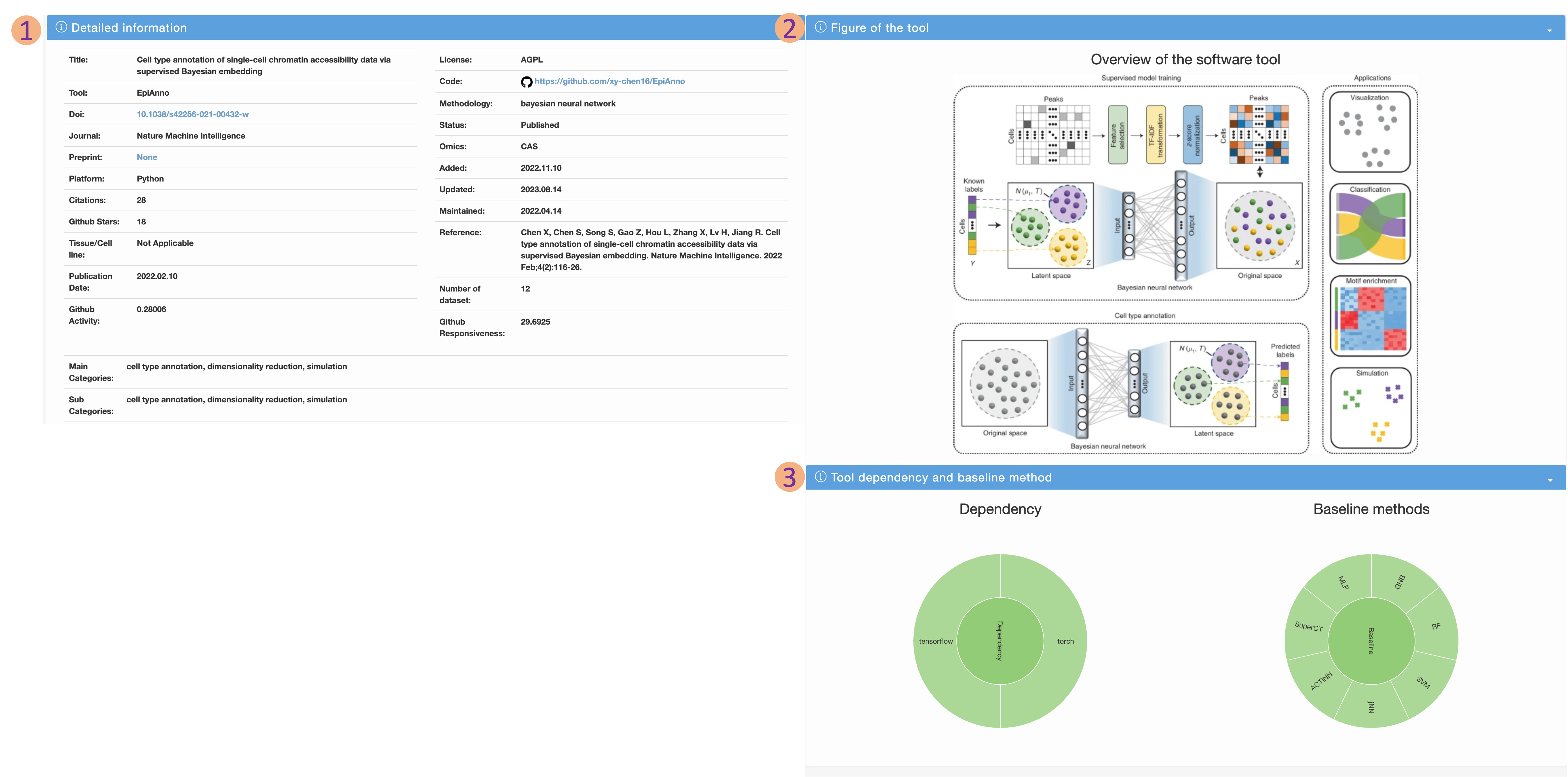
Details page
- Detailed information includes Doi, categories (main and sub), journal, platform, citations, stars, source code, methodology, omics, added time, updated time, code maintained time and baseline methods referred in the article.
- We also provide the figure of model or overview of each method. If the article does not propose a new model, we will record and display the first figure in the article. Users can intuitively understand the method according to this figure
- Besides, we also provide sunbrust figures showing the dependencies and baseline methods. Users can then compare this tool with the corresponding baseline methods, and also know the environment and tools they rely on to use this software tool
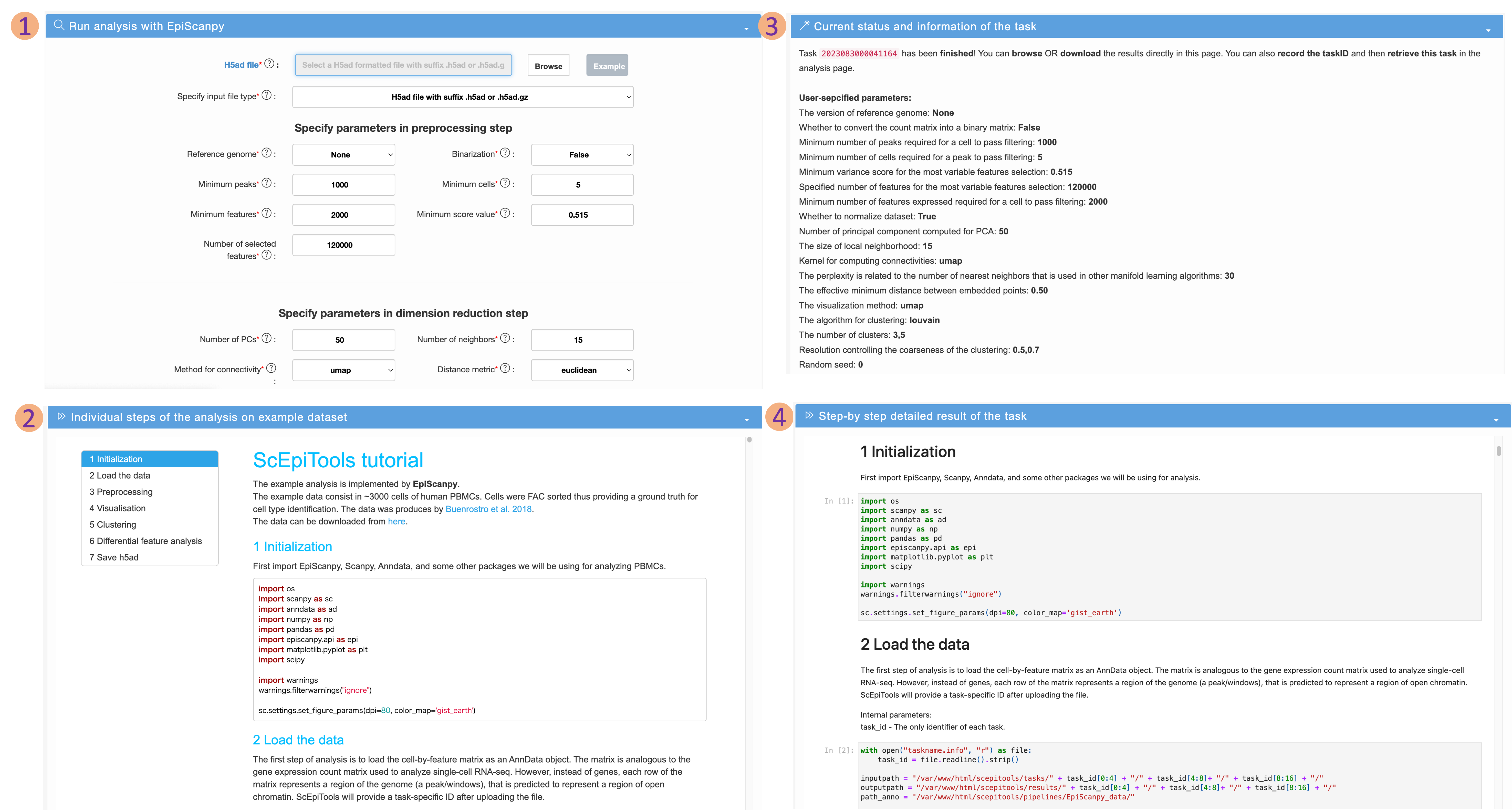
Analysis page
-
Users can select a kernel from the navigation bar to perform single-cell epigenomic data analysis. The usage process for the five different kernels is the same. Here, we take EpiScanpy as an example:
- Users can obtain the result of preprocessing, dimensionality reduction, visualization, clustering and differential chromatin analysis of single-cell chromatin accessibility sequencing data after submitting .h5ad-format file and setting parameters. User can also obtain a specific taskID for analysis results view and download.
- A demo of detailed steps of the analysis on example dataset.
- After submitting the input file and setting parameters, users can obtain the current status and information according to the taskID.
- Besides, users can view detailed analysis pipeline and results, including preprocessing, visualisation, clustering and differential feature analysis.
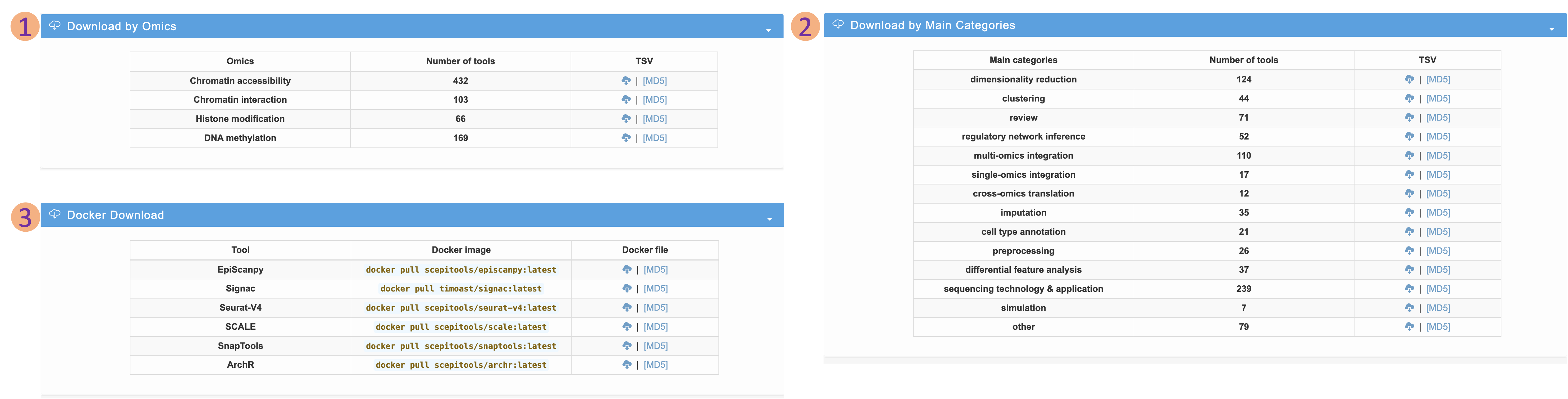
Download page
- We categorize the single-cell epigenomic tools by omics and categories to facilitate the data downloading.
- We also provide several docker files and docker images for the installation of environment of the software tools with the top recommendation scores
- The corresponding MD5 files are provided to ensure the integrity of the downloaded data.
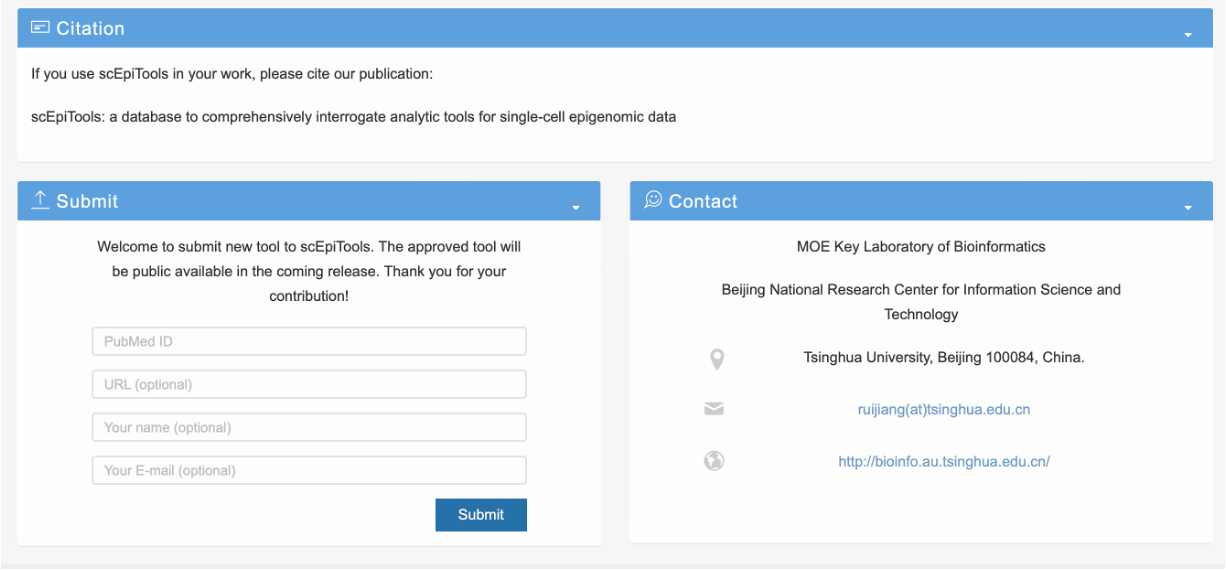
About page
- The citation information of scEpiTools.
- The interface for submitting data to scEpiTools. Users can submit their single-cell epigenetic software tool with the PubMed reference ID and optional information such as URL, name, and E-mail. The approved tool will be public available in the coming release. We appreciate the users for their contribution.
- The contact information of scEpiTools. Please feel free to contact us if you have any question about scEpiTools.
The browser compatibility of scEpiTools
| Operating System | Version | Chrome | Firefox | Opera | Microsoft Edge | Safari |
|---|---|---|---|---|---|---|
| Windows | Windows 10 Pro | |||||
| MacOS | Sierra 10.12 | Linux | Ubuntu 16.04 |
Frequently asked questions
What is ScEpiTools?
ScEpiTools, to the best of our knowledge, is the first comprehensive database for interrogating single-cell epigenomic tools, distinguished by its intuitive user interface, exhaustive data annotation, and extensive functional attributes. Epigenomic measures at single-cell resolution, particularly chromatin accessibility, DNA methylation, histone modifications, and chromosome conformation, have provided unprecedented opportunities to investigate the previously unknown heterogeneity of cell types, revolutionize the understanding of various complex tissues, and reveal a rich layer of regulatory information. The development of these new sequencing technologies has created a need for software tools to analyze single-cell chromatin data. Nonetheless, the overwhelming majority of effort has been focused on algorithm development, but this has not been fully investigated for systematic collection and task-specific recommendation of single-cell epigenomic tools. ScEpiTools aims to address this limitation and offers a promising avenue for researchers to investigate appropriate methods to analyze data. With systematic collection of 646 single-cell epigenomic tools, hierarchical categorization of usage, user-friendly search and recommendation, intuitive visualization, as well as detailed tutorial and demos of application, we anticipate that scEpiTools will benefit both biologists and data scientists for facilitating single-cell epigenomic data analysis, and empower them to achieve a better understanding of regulatory mechanisms.
What features ScEpiTools?
- Comprehensive collection of single-cell epigenetic tools.
- ScEpiTools is based on more detailed annotations.
- ScEpiTools hierarchically groups the tools into 14 major categories and 94 subcategories.
- Task-specific tool selection.
- Intuitive tutorial-style interfaces.
- Extensive application scenarios.
- ScEpiTools provides the first comprehensive collection of epigenomic tools, also with reviews, sequencing technologies, and studies. ScEpiTools now includes 646 articles, and is being constantly updated.
- ScEpiTools is based on more detailed annotations, each tool in scEpiTools was annotated with extensive details for recommendation, such as the tool methodology, baseline methods, dependency, etc.
-
ScEpiTools has organized single-cell epigenomic tools into 14 main task categories and 94 subcategories, which provides a comprehensive reference for researchers in this field. The main categories cover a wide range of tasks, including dimensionality reduction, clustering, regulatory network inference, and cell type annotation. In addition, ScEpiTools offers detailed subcategories for each main category, such as preprocessing tools which are subdivided into six categories, including reads alignment, quality control, data quality measure, peak calling, feature selection, and conversion from ArchR to Signac. By providing such a well-organized categorization, ScEpiTools facilitates the identification of suitable tools and streamlines the data analysis process for single-cell epigenomic studies.
- ScEpiTools enables task-specific tool selection via three novel recommendation scores, facilitating the analysis and benchmarking. The three recommendation scores are based on a weighted sum of different metrics such as impact, availability, time of publication, maintained status, and number of baseline methods and datasets used.
- ScEpiTools provides an intuitive tutorial-style interface, making it accessible to users without coding experience to perform single-cell epigenomic data analysis. The platform offers a complete analysis pipeline, including preprocessing, cell type annotation, and visualization, utilizing built-in kernels of epiScanpy (Danese et al., Nature Communications, 2021), Signac (Stuart et al., Nature Methods, 2021), SnapATAC (Fang et al., Nature Communications, 2021) and ArchR (Granja et al., Nature Genetics, 2021). By leveraging these mainstream tools as built-in kernels, scEpiTools can handle multiple formats of input data, improving its universality and usability. Morever, we have developed a novel ensemble kernel, called scEpiEnsemble, which demonstrates outstanding capacity in detecting epigenomic heterogeneity at the single-cell level. By markedly improving the precision of single-cell epigenomic data analysis, scEpiEnsemble facilitates a more comprehensive understanding of cell biology.
- ScEpiTools provides extensive application scenarios to further benefit both biologists and data scientists, such as advanced searching, detailed tutorial, data downloading by different groups. Additionally, scEpiTools provides Docker files and Docker images of several widely-used tools, which saves researchers time on environment installation.
Do you have any example for demonstrating the importance of single-cell epigentics software tools?
Single-cell epigenetic software tools are used routinely to identify new cell types, assess tissue composition, reveal regulatory mechanisms, and investigate cell development and lineage processes, via analyzing single-cell sequencing data of the epigenetic status.
For example, Zhang et al. applied snapATAC (Nature Communications 2021), a single nucleus analysis pipeline for ATAC-seq, to integrate > 1.3 million single-cell chromatin profiles from adult/fetal human tissues and create an atlas of ∼1.2 million candidate cis-regulatory elements across 222 cell types (Cell 2021). Among this atlas, they find the associations between cell types and noncoding variants of target genes in 240 complex human traits and diseases, and suggest relevant therapeutic targets in human cell types. Further use of this approach will help to identify cis-regulatory elements across distinct cell types infer potential key transcriptional regulators, which can control cell-type specific gene expression programs.
How to select suitable software tools for single cell epigenetic analysis?
ScEpiTools is a user-friendly database for efficently searching the analytical software tools of single-cell epigenomic data, users can select suitable software tools in the following two ways:
- via SEARCH page.
- via BROWSE page.
- Users can search for software tools of interest by selecting a platform, a category, an omics, a status, a journal, and a publication date from a series of pull-down menus, and or selecting multiple items from the pull-down menu with advanced features such as searching and selecting-all. Users can also determine the scope of the tool query by typing the need in the “Description” box. We also provide an example to facilitate the usage of searching.
The results will be displayed on an interactive table. Users can click the ID to access detailed information about this tool on a new page.
- Users can browse software tools of interest by selecting a main category and a subcategory in the hierarchical tree. We provide comprehensive statistics and visualization with respect to the selected subset of tools, and the selected subset of tools are displayed on a interactive table.
What do different Categories mean?
ScEpiTools grouped articles of single-cell epigenomic tools into 14 main categories. Each main-categories contain many subcategories, the detailed description of each main category is shown in the following table:
| Main Category | Description |
|---|---|
| dimensionality reduction | Projection of cells into a lower dimensional space |
| clustering | Unsupervised grouping of cells based on single-cell epigenomics profiles |
| review | Reviews covers computational methods, sequencing technologies, sequencing applications, and other relevant aspects for single-cell epigenomics |
| regulatory network inference | Identification or use of regulatory network between genes, peaks and DNA methylated regions |
| multi-omics integration | Integration of multi-omics datasets |
| single-omics integration | Combining of multiple single-omics datasets |
| cross-omics translation | Transformation between different omics datasets |
| imputation | Estimation of chromatin accessibility, DNA methylation, histone modification or chromatin interaction where zeros have been observed |
| cell type annotation | Assignment of cell types based on a reference dataset |
| preprocessing | Preprocessing for single-cell epigenomics datasets |
| differential feature analysis | Testing of differential feature across groups of cells |
| sequencing technology & application | Single-cell epigenomics sequencing technology or application |
| simulation | Generation of synthetic single-cell epigenomics datasets |
| other | Other analysis tasks for single-cell epigenomics data |
What is scEpiEnsemble kernel in the Analysis page?
According to statistics from ScEpiTools, dimensionality reduction is a crucial task in the analysis of single-cell epigenomic data and is necessary prior to downstream analyses such as visualization and clustering. In order to improve the quality of single-cell epigenomic data analysis, we have selected four methods with high recommendation scores and utilization, and have proposed scEpiEnsemble, an ensemble method designed to integrate the results of EpiScanpy, Signac, and SnapATAC. Through the ScEpiTools tutorial on the scEpiEnsemble kernel, we have validated the superior performance of scEpiEnsemble in clustering single-cell epigenomic data compared to three baseline methods.
How to prepare a h5ad file for online analysis?
H5ad-format file is a hdf5 file with additional structure and information specifying how to store AnnData objects. We have integrated options for users to upload files in the formats of 10x genomics, Seurat object, and SingleCellExperiment object on the Episcanpy and scEpiEnsemble pages. Users can upload data in these formats, and the server will automatically convert them into h5ad files before analysis. If users prefer to perform the data format conversion themselves or need to review the information within the h5ad file, please refer to the tutorial we have provided.